Shell Scripts
Overview
Teaching: 20 min
Exercises: 15 minQuestions
How can I save and re-use commands?
Objectives
Write a shell script that runs a command or series of commands for a fixed set of files.
Run a shell script from the command line.
Write a shell script that operates on a set of files defined by the user on the command line.
Demonstrate how to see what commands have recently been executed.
Create pipelines that include user-written shell scripts.
We are finally ready to see what makes the shell such a powerful programming environment. We are going to take the commands we repeat frequently and save them in files so that we can re-run all those operations again later by typing a single command. For historical reasons, a bunch of commands saved in a file is usually called a shell script, but make no mistake: these are actually small programs.
Our first shell script
Let’s start by going back to data and putting some commands into a new file called middle.sh using an editor like nano:
$ cd ~/shell-novice/data
$ nano middle.sh
So why the .sh extension to the filename? Adding .sh is the convention to show that this is a Bash shell script.
Now, enter the line head -15 sc_climate_data_1000.csv | tail -5 into our new file:
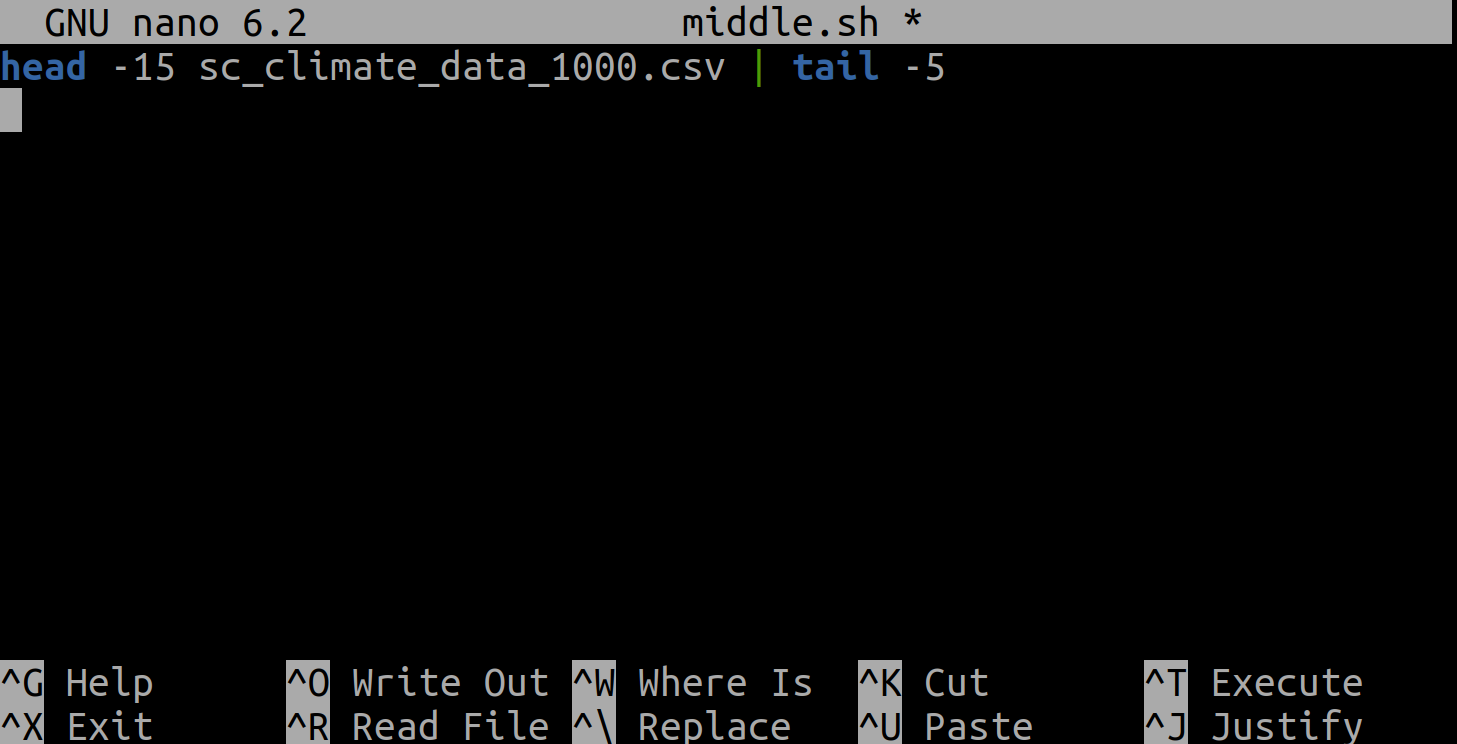
Then save it and exit nano (using Control-O to save it and then Control-X to exit nano).
This pipe selects lines 11-15 of the file sc_climate_data_1000.csv. It selects the first 15
lines of that file using head, then passes that to tail to show us only the last 5 lines - hence lines 11-15.
Remember, we are not running it as a command just yet:
we are putting the commands in a file.
Once we have saved the file,
we can ask the shell to execute the commands it contains.
Our shell is called bash, so we run the following command:
$ bash middle.sh
299196.8188,972890.0521,48.07,61.41,0.78
324196.8188,972890.0521,48.20,-9999.00,0.72
274196.8188,968890.0521,47.86,60.94,0.83
275196.8188,968890.0521,47.86,61.27,0.83
248196.8188,961890.0521,46.22,58.98,1.43
Sure enough, our script’s output is exactly what we would get if we ran that pipeline directly.
Text vs. Whatever
We usually call programs like Microsoft Word or LibreOffice Writer “text editors”, but we need to be a bit more careful when it comes to programming. By default, Microsoft Word uses
.docxfiles to store not only text, but also formatting information about fonts, headings, and so on. This extra information isn’t stored as characters, and doesn’t mean anything to tools likehead: they expect input files to contain nothing but the letters, digits, and punctuation on a standard computer keyboard. When editing programs, therefore, you must either use a plain text editor, or be careful to save files as plain text.
Enabling our script to run on any file
What if we want to select lines from an arbitrary file?
We could edit middle.sh each time to change the filename,
but that would probably take longer than just retyping the command.
Instead,
let’s edit middle.sh and replace sc_climate_data_1000.csv with a special variable called $1:
$ nano middle.sh
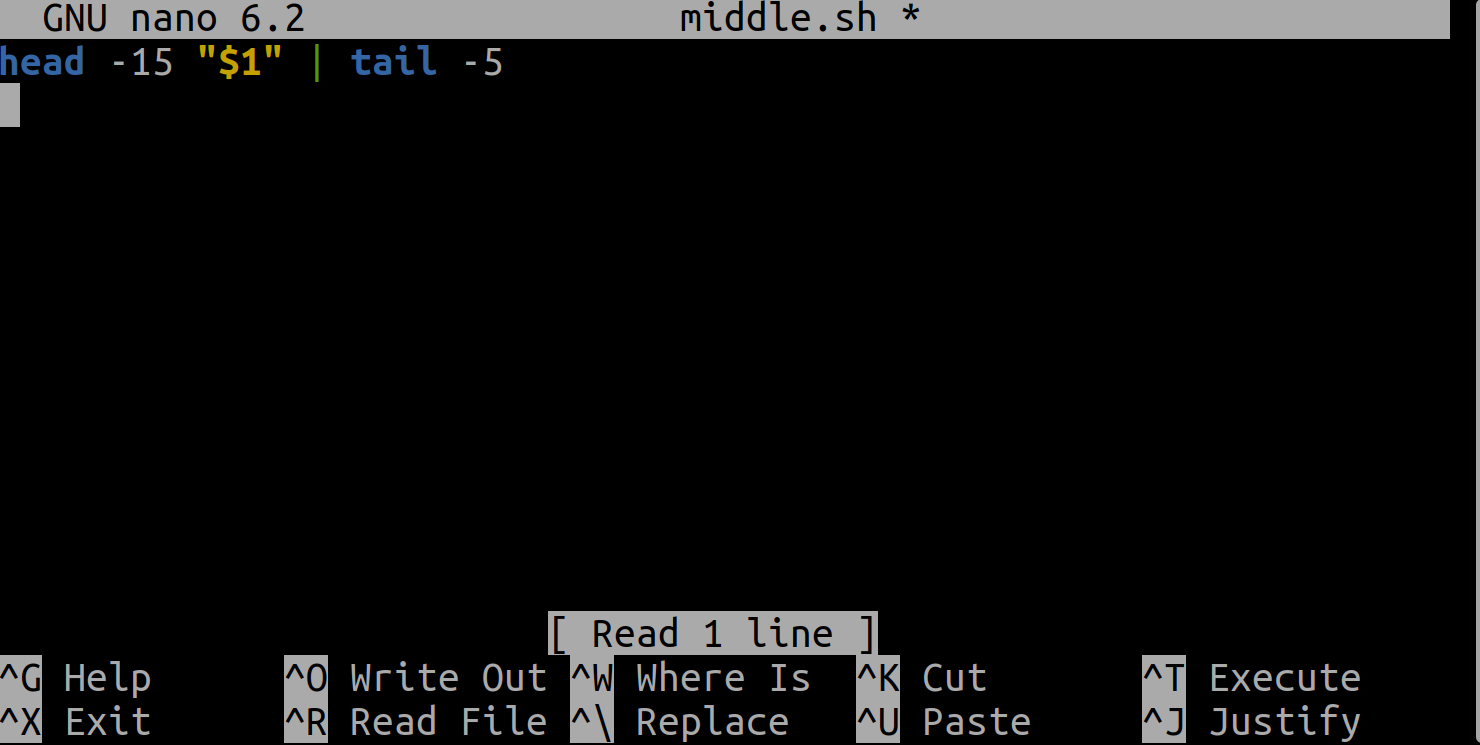
Inside a shell script,
$1 means the first filename (or other argument) passed to the script on the command line.
We can now run our script like this:
$ bash middle.sh sc_climate_data_1000.csv
299196.8188,972890.0521,48.07,61.41,0.78
324196.8188,972890.0521,48.20,-9999.00,0.72
274196.8188,968890.0521,47.86,60.94,0.83
275196.8188,968890.0521,47.86,61.27,0.83
248196.8188,961890.0521,46.22,58.98,1.43
or on a different file like this (our full data set!):
$ bash middle.sh sc_climate_data.csv
299196.8188,972890.0521,48.07,61.41,0.78
324196.8188,972890.0521,48.20,-9999.00,0.72
274196.8188,968890.0521,47.86,60.94,0.83
275196.8188,968890.0521,47.86,61.27,0.83
248196.8188,961890.0521,46.22,58.98,1.43
Note the output is the same, since our full data set contains the same first 1000 lines as sc_climate_data_1000.csv.
Double-Quotes Around Arguments
We put the
$1inside of double-quotes in case the filename happens to contain any spaces. The shell uses whitespace to separate arguments, so we have to be careful when using arguments that might have whitespace in them. If we left out these quotes, and$1expanded to a filename likeclimate data.csv, the command in the script would effectively be:head -15 climate data.csv | tail -5This would call
headon two separate files,climateanddata.csv, which is probably not what we intended.
Adding more arguments to our script
However, if we want to adjust the range of lines to extract, we still need to edit middle.sh each time.
Less than ideal!
Let’s fix that by using the special variables $2 and $3. These represent the second and third arguments passed on the command line:
$ nano middle.sh
Put the command line head "$2" "$1" | tail "$3" in the script. Now we can pass the head and tail line range arguments to our script:
$ bash middle.sh sc_climate_data_1000.csv -20 -5
252196.8188,961890.0521,46.22,60.94,1.43
152196.8188,960890.0521,48.81,-9999.00,1.08
148196.8188,959890.0521,48.81,59.43,1.08
325196.8188,957890.0521,48.20,61.36,0.72
326196.8188,957890.0521,47.44,61.36,0.80
This does work,
but it may take the next person who reads middle.sh a moment to figure out what it does.
We can improve our script by adding some comments at the top:
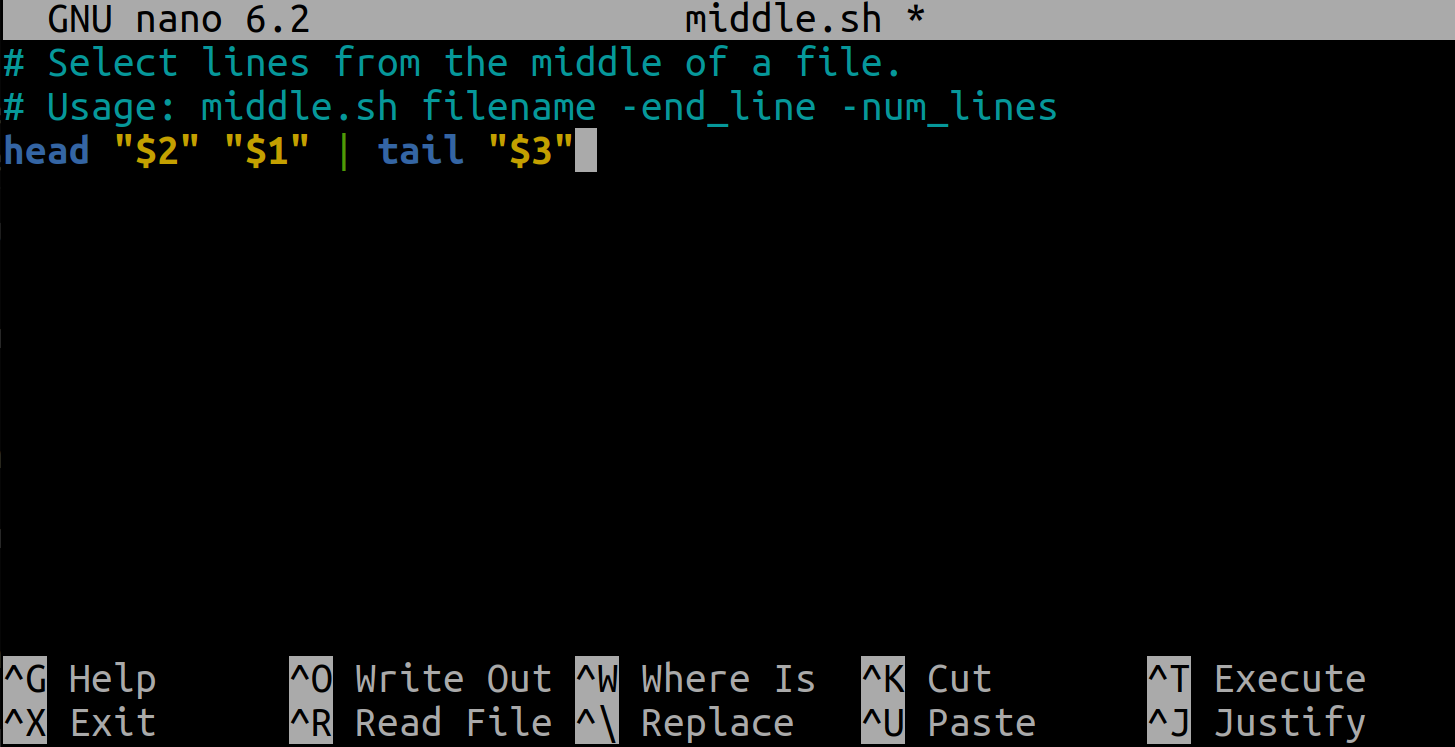
$ cat middle.sh
# Select lines from the middle of a file.
# Usage: middle.sh filename -end_line -num_lines
head "$2" "$1" | tail "$3"
In Bash, a comment starts with a # character and runs to the end of the line.
The computer ignores comments,
but they’re invaluable for helping people understand and use scripts.
A line or two of documentation like this make it much easier for other people (including your future self) to re-use your work. The only caveat is that each time you modify the script, you should check that its comments are still accurate: an explanation that sends the reader in the wrong direction is worse than none at all.
Processing multiple files
What if we want to process many files in a single pipeline?
For example, if we want to sort our .csv files by length, we would type:
$ wc -l *.csv | sort -n
This is because wc -l lists the number of lines in the files
(recall that wc stands for ‘word count’, adding the -l flag means ‘count lines’ instead)
and sort -n sorts things numerically.
We could put this in a file,
but then it would only ever sort a list of .csv files in the current directory.
If we want to be able to get a sorted list of other kinds of files,
we need a way to get all those names into the script.
We can’t use $1, $2, and so on
because we don’t know how many files there are.
Instead, we use the special variable $@,
which means,
“All of the command-line parameters to the shell script.”
We also should put $@ inside double-quotes
to handle the case of parameters containing spaces
("$@" is equivalent to "$1" "$2" …)
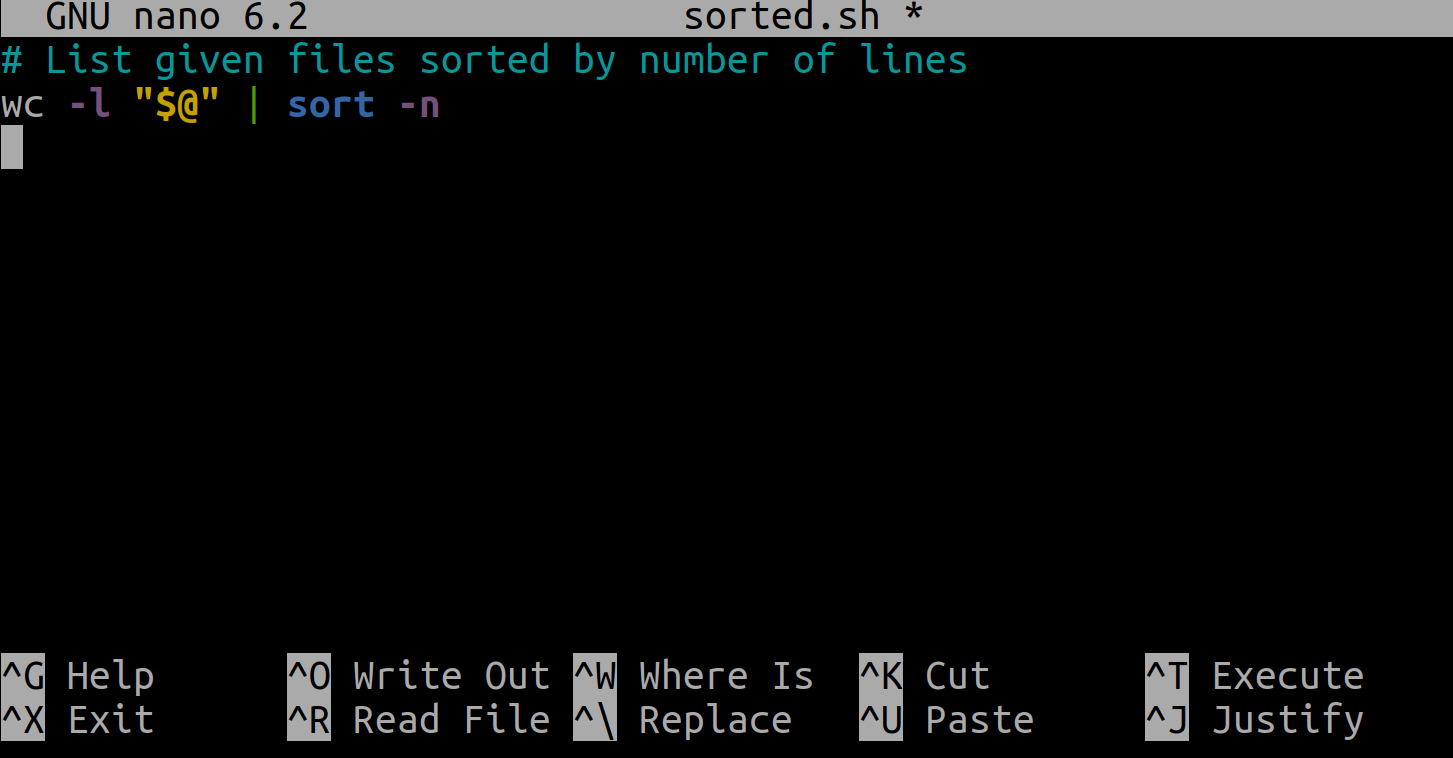
Here’s an example. Edit a new file called sorted.sh:
$ nano sorted.sh
And in that file enter:
wc -l "$@" | sort -n
When we run it with some wildcarded file arguments:
$ bash sorted.sh *.csv ../shell/test_directory/creatures/*.dat
We have the following output:
11 sc_climate_data_10.csv
155 ../shell/test_directory/creatures/minotaur.dat
163 ../shell/test_directory/creatures/basilisk.dat
163 ../shell/test_directory/creatures/unicorn.dat
1001 sc_climate_data_1000.csv
1048576 sc_climate_data.csv
1050069 total
Why Isn’t It Doing Anything?
What happens if a script is supposed to process a bunch of files, but we don’t give it any filenames? For example, what if we type:
$ bash sorted.shbut don’t say
*.dat(or anything else)? In this case,$@expands to nothing at all, so the pipeline inside the script is effectively:wc -l | sort -nSince it doesn’t have any filenames,
wcassumes it is supposed to process standard input, so it just sits there and waits for us to give it some data interactively. From the outside, though, all we see is it sitting there: the script doesn’t appear to do anything.If you find yourself in this situation pressing
Control-Cwill stop the command from taking input and return you to the command line prompt.Again, we should explain what we are trying to do here using a comment, for example:
What did I type to get that to work?
Here’s something that can be useful as an aid to memory. Suppose we have just run a series of commands that did something useful. For example, that created a graph we’d like to use in a paper. We’d like to be able to re-create the graph later if we need to, so we want to save the commands in a file. Instead of typing them in again (and potentially getting them wrong) we can do this:
$ history | tail -4 > redo-figure-3.shThe file
redo-figure-3.shnow contains:$ cat redo-figure-3.sh297 bash goostats NENE01729B.txt stats-NENE01729B.txt 298 bash goodiff stats-NENE01729B.txt /data/validated/01729.txt > 01729-differences.txt 299 cut -d ',' -f 2-3 01729-differences.txt > 01729-time-series.txt 300 ygraph --format scatter --color bw --borders none 01729-time-series.txt figure-3.pngAfter a moment’s work in an editor to remove the historical reference number for each command (e.g. 297, 298), we have a completely accurate record of how we created that figure. Note that to use the commands in the history simply type its related reference number after
!in the terminal (e.g, type!297to re-rungoostatsonNENE01729B.txt)Other History Commands
There are a number of other shortcut commands for getting at the history.
Ctrl-Renters a history search mode “reverse-i-search” and finds the most recent command in your history that matches the text you enter next. PressCtrl-Rone or more additional times to search for earlier matches.!!retrieves the immediately preceding command (you may or may not find this more convenient than using the up-arrow)!$retrieves the last word of the last command. That’s useful more often than you might expect: afterbash goostats NENE01729B.txt stats-NENE01729B.txt, you can typeless !$to look at the filestats-NENE01729B.txt, which is quicker than doing up-arrow and editing the command-line.In practice, most people develop shell scripts by running commands at the shell prompt a few times to make sure they’re doing the right thing, then saving them in a file for re-use. This style of work allows people to recycle what they discover about their data and their workflow with one call to
historyand a bit of editing to clean up the output and save it as a shell script.
Exercises
Variables in shell scripts
In the
~/shell-novice/shell/test_directory/moleculesdirectory, you have a shell script calledscript.shcontaining the following commands:$ cat script.shhead $2 $1 tail -n $3 $1Note that here, we use the explicit
-nflag to pass the number of lines totailthat we want to extract, since we’re passing in multiple.pdbfiles. Otherwise,tailcan give us an error about incorrect options on certain machines if we don’t.While you are in the molecules directory, you type the following command:
bash script.sh '*.pdb' -1 -1Which of the following outputs would you expect to see?
- All of the lines between the first and the last lines of each file ending in
*.pdbin the molecules directory- The first and the last line of each file ending in
*.pdbin the molecules directory- The first and the last line of each file in the molecules directory
- An error because of the quotes around
*.pdbSolution
The answer is 2. The quotes around the wildcard
'*.pdb'mean it isn’t expanded when we call the script - but it will get expanded inside the script. There, it gets expanded to match every file in the directory that ends in*.pdb, and effectively the script calls:head -1 *.pdb tail -n -1 *.pdb*This prints out the first line (
head -1) of each.pdbfile, and then the last line of each.pdbfile.If we’d called the script as:
bash script.sh *.pdb -1 -1Then it wouldn’t work as the wildcard would’ve expanded before the script started and we’d have effectively run it as:
bash script cubane.pdb ethane.pdb methane.pdb octane.pdb pentane.pdb propane.pdb -1 -1This would have caused an error, as we expect the second and third arguments to be numbers for
headandtail!
Script reading comprehension
Joel’s
datadirectory contains three files:fructose.dat,glucose.dat, andsucrose.dat. Explain what a script calledexample.shwould do when run asbash example.sh *.datif it contained the following lines:# Script 1 echo *.*# Script 2 for filename in $1 $2 $3 do cat $filename done# Script 3 echo $@.datSolution
Script 1 doesn’t use any arguments - so it ignores our
*.daton the command line. The*.*wildcard matches anything in the current directory with a.in the file (or folder!) name, so it expands to a list of all files in the directory, includingexample.sh. Then it passes that list toecho, which prints them out.example.sh fructose.dat glucose.dat sucrose.datScript 2 makes use of our arguments. The wildcard
*.datmatches any file that ends in.dat, so expands tofructose.dat glucose.dat sucrose.datthen passes them to the script. The script then takes the first 3 arguments (using$1 $2 $3) and usescatto print the contents of the file. However, if there are more than 3 files in the directory with the.datsuffix, they’ll be ignored. If there are less than 3, there’ll be an error!Script 3 uses all our arguments - the
$@variable gets expanded into the full list of arguments,fructose.dat glucose.dat sucrose.dat.echothen prints out that list… with.datadded to the end of it:fructose.dat glucose.dat sucrose.dat.datThis probably isn’t quite what we were hoping for!
Key Points
Save commands in files (usually called shell scripts) for re-use.
bash [filename]runs the commands saved in a file.
$@refers to all of a shell script’s command-line arguments.
$1,$2, etc., refer to the first command-line argument, the second command-line argument, etc.Use Ctrl+R to search through the previously entered commands.
Use
historyto display recent commands, and![number]to repeat a command by number.Place variables in quotes if the values might have spaces in them.
Letting users decide what files to process is more flexible and more consistent with built-in Unix commands.